Daniel McNeela
Machine Learning Researcher and Engineer
Arizona State University
University of Wisconsin, Madison
Biography
Hi! I am a machine learning engineer and researcher, a bioinformatician, a professional content writer, and a law student. I am currently pursuing a JD at Arizona State University’s Sandra Day O’Connor College of Law and a PhD in Computer Science from the University of Wisconsin, Madison. I earned an M.S. in Biomedical Data Science from the University of Wisconsin, Madison and a B.A. in Applied Mathematics from the University of California, Berkeley. During my M.S., I was lucky to be advised by Professors Anthony Gitter and Fred Sala, both of whom have had an outsized impact on my development as a researcher. My current academic research focuses on applying tools from geometric deep learning to computational chemistry and biology problems such as drug discovery, equivariance, machine learning interatomic potentials, and multiomics. On the law side, I am interested in the intersection of law and technology, particularly patent and intellectual property law surrounding generative AI as well as AI regulation and its policy implications.
In addition to my academic interests, I enjoy running, cycling, Brazilian jiu-jitsu, playing guitar, piano, and drums, writing creative fiction, and reading great books. In particular, writing is a huge passion of mine. I work part-time as a professional content and technical writer specializing in machine learning, so please contact me if you are looking for a skilled writer for your software or ML business.
- Patent Law
- IP & Copyright Law
- Machine Learning
- Geometric Deep Learning
- Equivariance
- Computational Biology
Juris Doctor (JD), 2027
Arizona State University
PhD in Computer Science, On Pause
University of Wisconsin, Madison
MS in Biomedical Data Science, 2023
University of Wisconsin, Madison
BA in Applied Mathematics, 2017
University of California, Berkeley
Recent Posts
Featured Publications
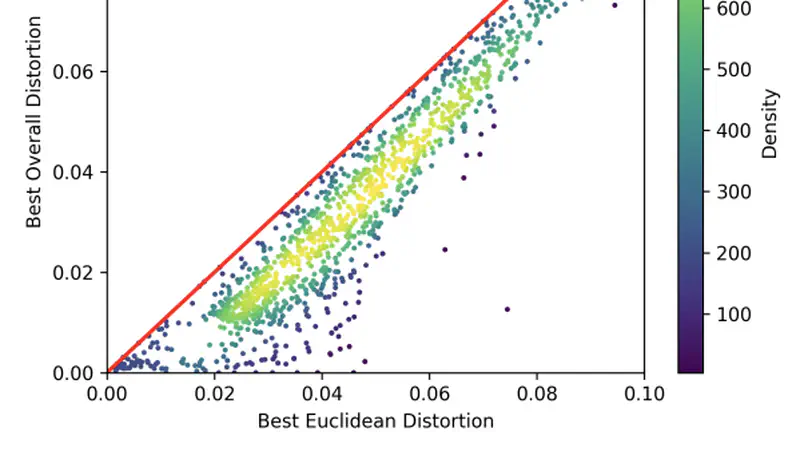
Machine learning models that embed graphs in non-Euclidean spaces have shown substantial benefits in a variety of contexts, but their application has not been studied extensively in the biological domain, particularly with respect to biological pathway graphs. Such graphs exhibit a variety of complex network structures, presenting challenges to existing embedding approaches. Learning high-quality embeddings for biological pathway graphs is important for researchers looking to understand the underpinnings of disease and train high-quality predictive models on these networks. In this work, we investigate the effects of embedding pathway graphs in non-Euclidean mixed-curvature spaces and compare against traditional Euclidean graph representation learning models. We then train a supervised model using the learned node embeddings to predict missing protein-protein interactions in pathway graphs. We find large reductions in distortion and boosts on in-distribution edge prediction performance as a result of using mixed-curvature embeddings and their corresponding graph neural network models. However, we find that mixed-curvature representations underperform existing baselines on out-of-distribution edge prediction performance suggesting that these representations may overfit to the training graph topology. We provide our Mixed-Curvature Product Graph Convolutional Network code at this https URL and our pathway analysis code at this https URL.
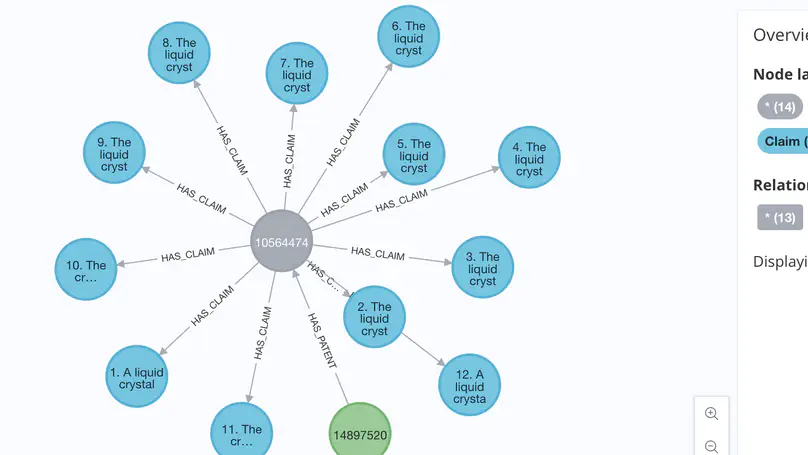
In this work we introduce Graph Retrieval-Optimized Generation (GROG), a method for reducing LLM hallucinations in contexts where external, graph-structured knowledge is available. We test our method on retrieval and generation tasks conditioned on publicly-available USPTO patent data and show promising results, suggesting that this method warrants further study in more diverse legal contexts and downstream applications.
Recent Publications
Experience
Contact
- mcneelaml@gmail.com
- Washington, DC 20037
- DM Me