The Variant Calling Problem
In bioinformatics, particularly in the subfield of oncology in which I work, we’re often tasked with the issue of identifying variants in a genomic sequence. What this means is that we have a sample sequence along with a reference sequence, and we want to identify regions where the sample differs from the reference. These regions are called variants, and they can often be clinically relevant, potentially indicating an oncogenic mutation or some other telling clinical marker.
In general, there are two types of variant calling that one may be interested in performing. The first of these is called germline variant calling. In this type, we use a reference genome that is an accepted standard for the species in question. There exists more than one acceptable choice for a reference genome, but one of the current standards for humans is called GRCh38. This stands for Genome Reference Consortium human and 38 is an identifying version number. Note the h is used to distinguish the human genome from other genomes that the GRC puts out such as GRCm (mouse genome).
The second type is called somatic variant calling. This method uses two samples from a single individual and compares the sequence of one with the other. In effect, the patients own genome serves as the reference. This is the method commonly used in oncology, as one sample can be taken from a tumor and another from some non-mutated cell such as the bloodline. This process is also sometimes called a tumor-normal study due to the fact that one sample is taken from the tumor and one from a normal cell.
There are a few additional things to note in the discussion above. The first is that in germline calling we’re uncovering variants that are being passed from parent to child via the germ cells, i.e. the sperm and eggs. In somatic calling, we’re identifying somatic mutations and variants, those that occur within the body over the course of a lifetime, whether they are due to environmental factors, errors in DNA transcription and translation, or some other factor.
Types of Variants
In order to identify variants, it will be helpful to develop an ontology of some of the specific classes and types of variants we might expect to see as the output of our variant callers.
Indel - This is one of the simplest types of variant to describe conceptually, but also one of the most difficult to identify via current variant calling methods. An indel is either an insertion or deletion of a single base at some point in the DNA sequence. The reason these are difficult to identify is that since it is not known in advance at which point in the DNA the variant occurs, it can throw off the alignment with the reference sequence.
SNP - This is short for single nucleotide polymorphism and it refers to a population-level variant in which a single base differs between the sample and reference sequences. By population-level, we mean that this variant is extremely common across specific populations. For example, an SNP might occur when comparing the genomes of Caucasian and Chinese people within a gene coding for skin tone.
SNV - This stands for single-nucleotide variant and is the same as an SNP apart from the fact that it indicates a novel mutation present in the genomes of a few rather than being widespread across a population.
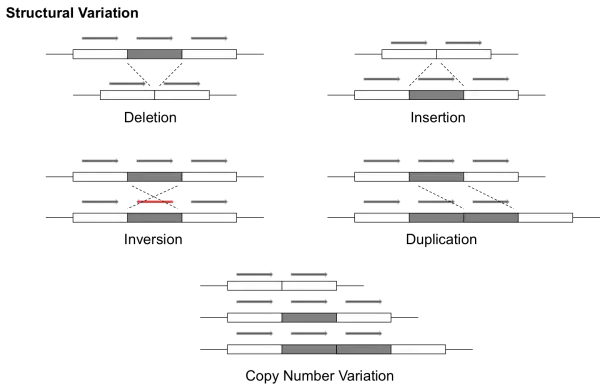
Structural Variant - A structural variant refers to a larger-scale variant in the genome, typically occupying at least 1000 base pairs. These can exhibit a number of different behaviors. For example, a >1kb region of the DNA can be duplicated and reinserted or it can be deleted. These types of structural variation are commonly referred to as copy number variants (CNVs). A partial sequence can also be reversed (called inversion). This image sums it up well
Haplotype Phasing
Before getting into the nitty gritty of variant calling, it will be helpful to describe a related process called haplotype phasing. A haplotype is the set of genetic information associated with a single chromosome. The human genome is diploid, meaning it consists of pairs of chromosomes for which each has one part of its genetic information inherited from the mother and the other part from the father. The combined genetic information resulting from considering these pairs of chromosomes as single units is called the genotype. However, having access to the haplotype can be crucial because it can provide information crucial to identifying disease-causing variants (either SNPs or SNVs) in the genome. The issue is that current sequencing methods don’t give us haplotype information for free as often the reads produced cannot be separated into their individual male and female loci. Therefore, we need to use statistical algorithms to piece together what we can about the haplotype sequences after the fact. This is where haplotype phasing comes into play.
Some Preliminaries and Associated Challenges
To start with, we need to establish some background with regards to population-level frequencies of genetic variation so that we can begin to detect deviations from that standard. What may be surprising is that while the human genome is diploid, it varies from the reference at approximately 0.1% of the bases. That means that between any two individuals, we should expect to see at least 99.9% shared genetic information. It is at the remaining 0.1% of sites that we direct our attention when we’re looking to tease out the haplotypes. For reference, the condition of sharing genetic information is often called homology.
The teasing out of haplotypes is complicated by the fact that at times genetic information is passed from parent to child in a process known as recombination. Recombination happens when instead of passing down just one chromosome from its diploid pair, a parent passes down a combination of the two. At the biological level, this process occurs during meiosis. The first phase of meiosis involves an alignment of homologous chromosome pairs between the mother and father. This process involves a stage in which the chromosomal arms temporarily overlap, causing a crossover which may (or may not) result in fusion at that locus at which the point of crossover occurs. This results in a recombination of genetic information which is then passed down to the child. One benefit to this process is that it encourages genetic diversity, creating genetic patterns in the child that appeared in neither parent. Unfortunately, it also makes haplotype phasing that much more difficult.
DNA sequencing gives partial haplotype information. It produces sequencing reads which are partial strings of the sequence up to 1000 base pairs. These reads all come from the same chromosome, but they represent only a small slice of the haplotype because the entire chromosome is on the order of 50 million to 250 million base pairs.
Advantages of Knowing the Haplotype
Hopefully it’s clear that having access to the haplotype for any given individual is strictly superior to simply having the genotype. This is because, given access to a person’s haplotype, we can simply combine that information in a procedural way to yield their genotype. The same cannot be said of the reverse. What’s more, having the haplotype allows us to compute statistics about various biological properties that would otherwise be unavailable to use. For example, if we have haplotypes for a series of individuals in a population, we can check at which loci recombination is most likely to occur, what the frequency of that recombination is, etc. It suffices to say that having access to the haplotype is highly advantageous as a downstream input to variant calling methods, and I’ll examine some such methods that make use of this information in upcoming posts.
The DeepVariant Caller
Now that we have an understanding of haplotypes and some of the different ways in which variants might arise, I’d like to introduce one of the most successful models for variant calling created to-date: Google’s DeepVariant. This model is one of the simplest to introduce in that it uses virtually no specialized knowledge of genomics in terms of encoding input features. Surprisingly, this has no detrimental effect on its accuracy, as it outperformed virtually every other variant caller on the market at the time of its release. The approach it uses is somewhat novel so I’ll introduce that here, and I’ll compare it in subsequent posts with the methodology behind other variant callers.
How it Works
DeepVariant is unique in that it doesn’t operate on the textual sequence information of the aligned and reference genomic reads. Rather, it creates something called a pileup image from the alignment information. A pileup image shows the sequenced reads from the sample aligned with the reference. Each base is assigned an RGB color, and possible variants within the reads as compared to the reference are highlighted accordingly. Here’s an example of what a pileup looks like.
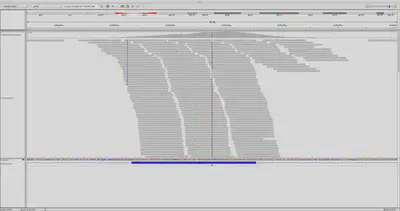
The DeepVariant model is trained on a large dataset of such images in which variants have already been labeled. It can then be applied to new alignments and samples by generating their pileup using a program such as SAMTOOLS. Here’s how the authors from Google structured their model.
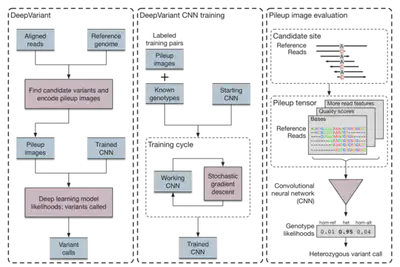
As you can see from the diagram, their workflow proceeds as follows:
- Identifying Candidate Variants and Generating Pileup: Candidate variants are identified according to a simple procedure. The reads are aligned to the reference, and in each case the CIGAR string for the read is generated. Based on how that read compares to the reference at the point which it is aligned, it is classified as either a match, an SNV, an insertion, or a deletion. If the number of reads which differ at that base pass a pre-defined threshold, then that site is identified as a potential variant. The authors also apply some preprocessing to ensure that the reads under consideration are of high enough quality and are aligned properly in order to be considered. After candidate variants have been identified, the pileup image is generated. The notable points here are that candidate variants are colored and areas with excessive coverage are downsampled prior to image generation.
- The model is trained on the data, then run during inference stages. They use a CNN, specifically the Inception v2 architecture, which takes in a 299x299 input pileup image and uses an output softmax layer to classify into one of hom-ref, het, or hom-alt. They use SGD to train with a batch size of 32 and RMS decay set to 0.9. They initialize the CNN to using the weights from one of the ImageNet models.
The authors find that their model generalizes well, and can be used with no statistically significant loss in accuracy on reads aligned to a reference different from the one on which DeepVariant was trained. They also stumble upon another surprising result, namely that their model successfully calls variants on the mouse genome, opening up the model’s to a wide diversity of species.
Conclusion
I hope this post shed some light on how variant calling works and elucidates the biological underpinnings of the process well enough for newcomers to the field to start diving into some papers. In future posts, I’ll expand on some of the other popular variant calling models as well as introduce algorithms for related processes, such as the haplotype phasing discussed earlier.